Pre-Processing of microscopic volumetric data
Introduction
Most image analysis problems in the microscopic field can only be solved, when the whole processing pipeline is considered and optimized. This pipeline starts at the interaction of light with the biological sample, includes the optical transformation of the wave field, the conversion of photons to electrons, the electronic processing and finally the analog to digital conversion of the obtained charges.
Therefore we regard the 3D image recording process as the process of measuring certain physical quantities (e.g. the local concentration of a fluorecent marker) with the required spatial, temporal, and dynamic resolution.
The goal of the image pre-processing is therefore to eliminate or reduce known errors and shortcomings of the optical and electronic system. This could be achieved by introducing additional prior knowledge (e.g. an image formation model and the transfer functions of the optical system), or by using multiple recordings of the same object with varying recording parameters.
Multiview fusion of microscopical images
Members: Maja Temerinac-Ott , Dr. Olaf Ronneberger, Prof. Dr. Hans BurkhardtPartners: Dr. Roland Nitschke ( Centre of Systems Biology (ZBSA)), Prof. Dr. Wolfgang Driever
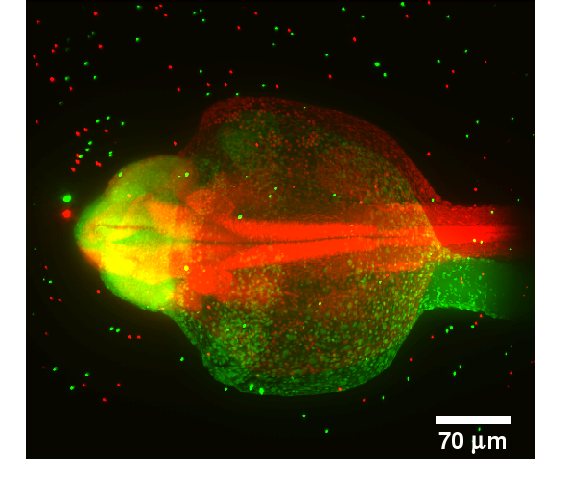
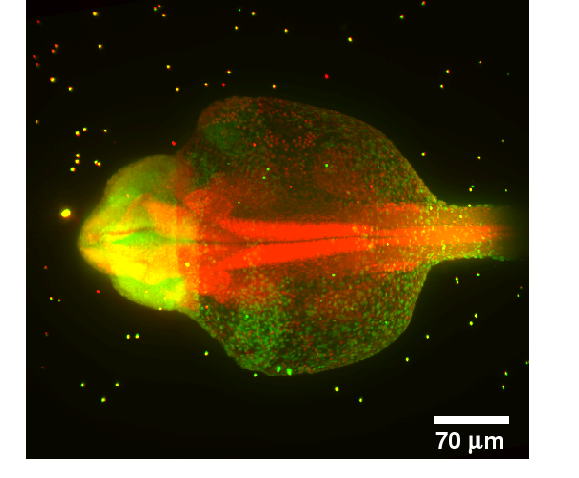
IntroductionWhen imaging an object in three dimensions, the resolution in axial direction is always smaller than the resolution in xy-direction. As the imaging gets deeper into the object, light gets absorbed. The missing information can be obtained by transforming the missing part of the object to the imaging plane, thus resulting in multiple images of the same object. The Single Plane Illumination Microscopy (SPIM) can produce such multiview images.GoalsThe goal of this project is the development of fast and reliable algorithms for multiview reconstruction of microscopical images for recording of large specimens (e.g. zebrafish embryos).ApproachThe reconstruction is decomposed in two steps: Registration and Fusion. For the registration step we assume that beads have been inserted into the probe and therefore the bead positions are used for the registration step. The registration is decomposed into three steps. 1.Extraction of the bead positions using morphological operations 2.Finding initial correspondences between beads using a Local Descriptor 3.Estimation of the transformation for registration using the found correspondences and refinement of the final transformation The image fusion is formulated as a multi view deconvolution problem based on the image formation model. An iterative algorithm is developed in order to find the best estimate of the original image based on the structural similarity index measure (SSIM).AchievementsA robust point cloud local descriptor based on Group Averaging for registration of the beads was developed [2] and applied to Single Plane Illumination Microscopy (SPIM) images. The computation of the final transformation between six views with 100 beads per view is performed in 10 seconds. A new fusion algorithm based on the SSIM index was developed [1] and successfully applied to artificially modelled two dimensional multi view images.Publications
|
|
Stitching of large 3D data sets
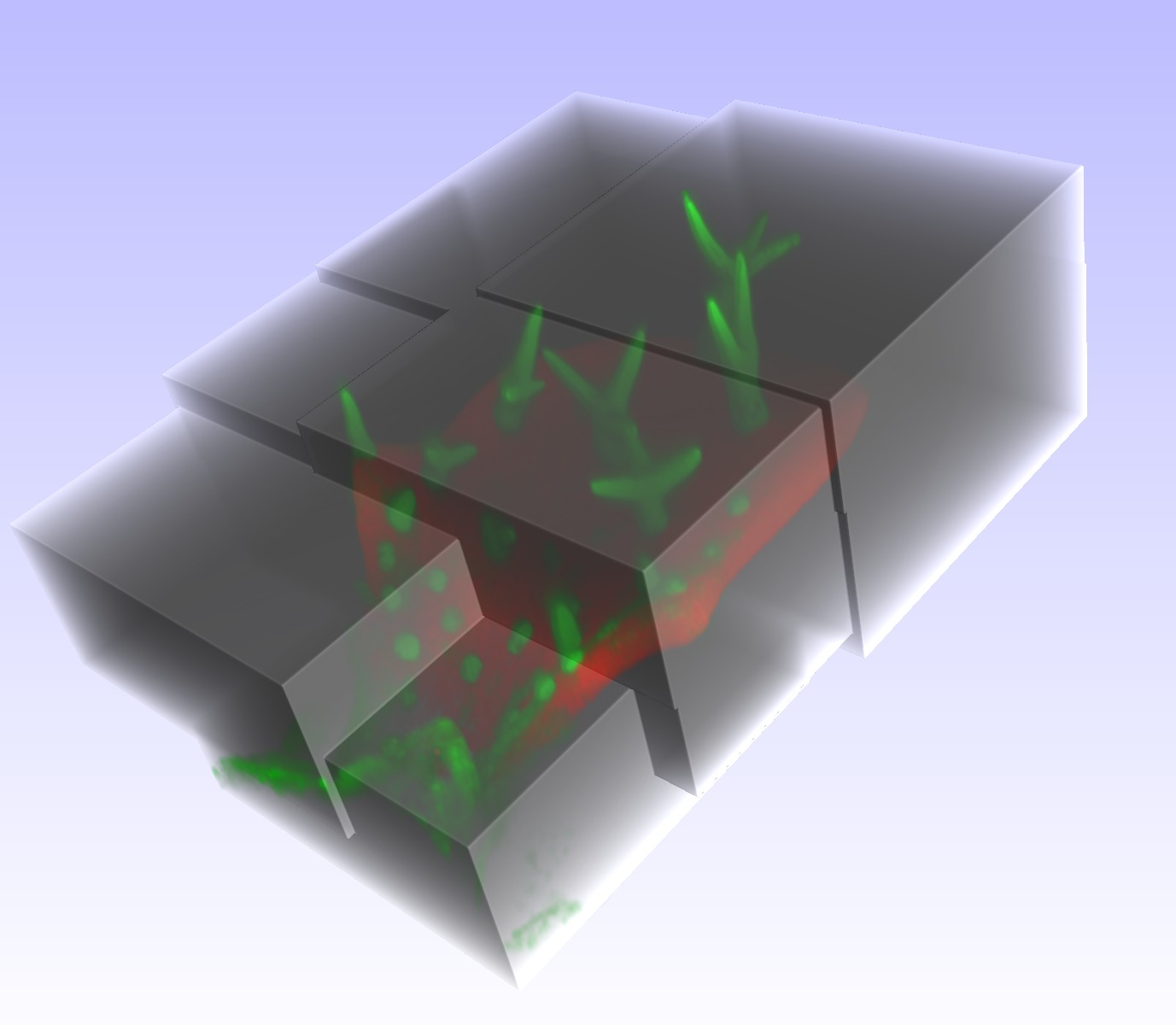
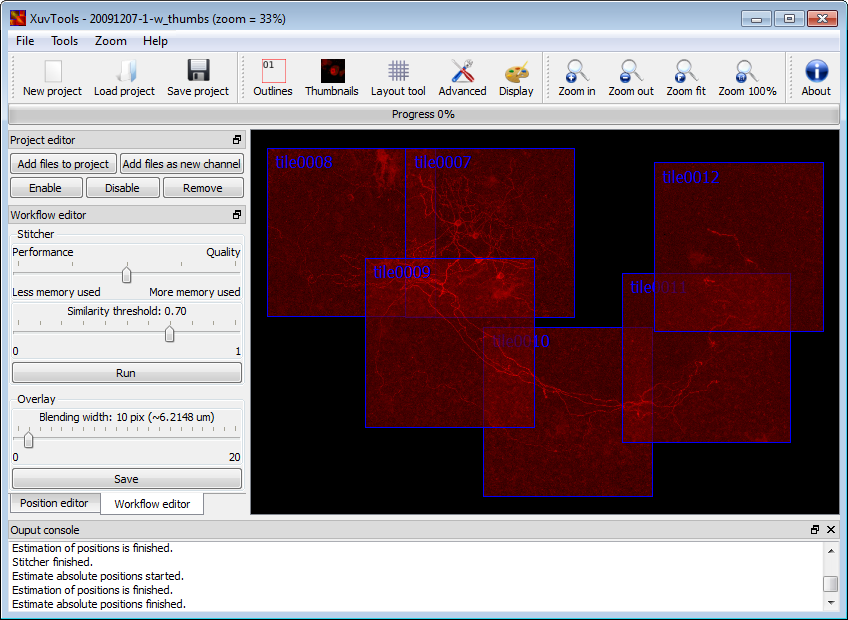
IntroductionCurrent biomedical research increasingly requires imaging large and thick 3D structures at high resolution. Prominent examples are the tracking of fine filaments over long distances in brain slices, or the localization of gene expression or cell migration in whole animals like Caenorhabditis elegans or zebrafish. To obtain both high resolution and a large field of view (FOV), a combination of multiple recordings ("tiles") is one of the options. Although hardware solutions exist for fast and reproducible acquisition of multiple 3D tiles, generic software solutions aremissing to assemble ("stitch") these tiles quickly and accurately.In this paper, we present a framework that achieves fully automated recombination of tiles recorded at arbitrary positions in 3D space, as long as some small overlap between tiles is provided. A fully automated 3D correlation between all tiles is achieved such that no manual interaction or prior knowledge about tile positions is needed. We use (1) phase only correlation in a multi-scale approach to estimate the coarse positions, (2) normalized cross-correlation of small patches extracted at salient points to obtain the precise matches, (3) find the globally optimal placement for all tiles by a singular value decomposition and (4) accomplish a nearly seamless stitching by a bleaching correction at the tile borders. If the dataset contains multiple channels, all channels are used to obtain the best matches between tiles. For speedup we employ a heuristic method to prune unneeded correlations, and compute all correlations via the fast Fourier transform (FFT), thereby achieving very good runtime performance. We demonstrate the successful application of the proposed framework to a wide range of different datasets from whole zebrafish embryos and C. elegans, mouse and rat brain slices and fine plant hairs (trichome). Further, we compare our stitching results to those of other commercially and freely available software solutions. The algorithms presented are being made available freely as an open source toolset "XuvTools" at http://www.xuvtools.org/ licensed under the GNU General Public License (GPL) v2. Binaries are provided for Linux and Microsoft Windows. The toolset is written in templated C++, such that it can operate on datasets with any bit-depth. Due to the consequent use of 64bit addressing, stacks of arbitrary size (i.e. larger than 4 GB) can be stitched. The runtime on a standard desktop computer is in the range of a few minutes. A user friendly interface for advanced manual interaction and visualization is also available. |
|
||
Publications
|
|||
This page is maintained by the project responsibles.


